
2017年3月14日,國際期刊《Biotechnology and Bioengineering》上在線發表了華東理工大學國家重點實驗室、國家生化工程技術研究中心(上海)儲炬教授課題組題為”A 9-pool metabolic structured kinetic model describing days to seconds dynamics of growth and product formation by Penicillium chrysogenum” 的研究論文。上海國佳生化工程技術研究中心有限公司項目負責人唐文俊為論文第一作者,儲炬教授為論文通訊作者。相關研究為與荷蘭DSM集團及荷蘭代爾夫特理工大學共同開展。
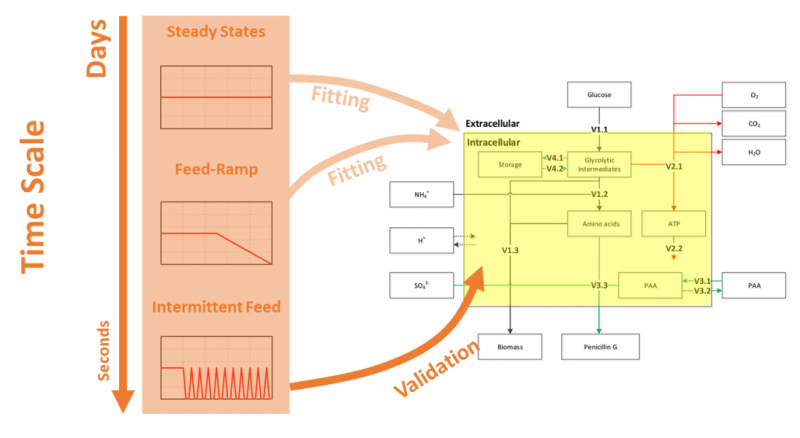
青霉素的生物過程優化已經有了幾十年的歷史,其工業發酵規模已經達到數百噸,效價也已從最初的幾十幾百U/ml優化只十幾萬U/ml。然而,通過系統地生物學研究發現,當前的產能甚至不足菌株最大產能的50%,依舊具有較大的優化空間。更為重要的是,工業化的青霉素生產往往還會遇到實驗室研究中不曾考慮的問題,其中最為主要的問題便是由于規模的增大導致噸級反應器內底物、溶氧、能量等不均一分布的出現,這也是生物過程工業放大所面臨的最大問題。基于上述背景,國家生化工程技術研究中心(上海)與著名跨國生物企業荷蘭DSM集團及國際一流院校荷蘭代爾夫特理工大學共同開展基于理性工業放大的好氧發酵生物過程研究。以產黃青霉作為研究對象,通過建立與細胞生理學相關的細胞反應動力學模型(CRD)以及與生物反應器流場相關的計算流體力學計算框架(CFD),并將兩者結合(CFD+CRD)來實現對于大型生物反應器的全尺寸模擬,并基于定量的模擬結果實現針對性的工業放大優化策略制定。該技術路線的難點便是建立相關的細胞動力學模型,該模型需要具備模型結構相對簡單,模型魯棒性強,動態過程模擬、秒至小時級細胞動力學特征的捕捉等。
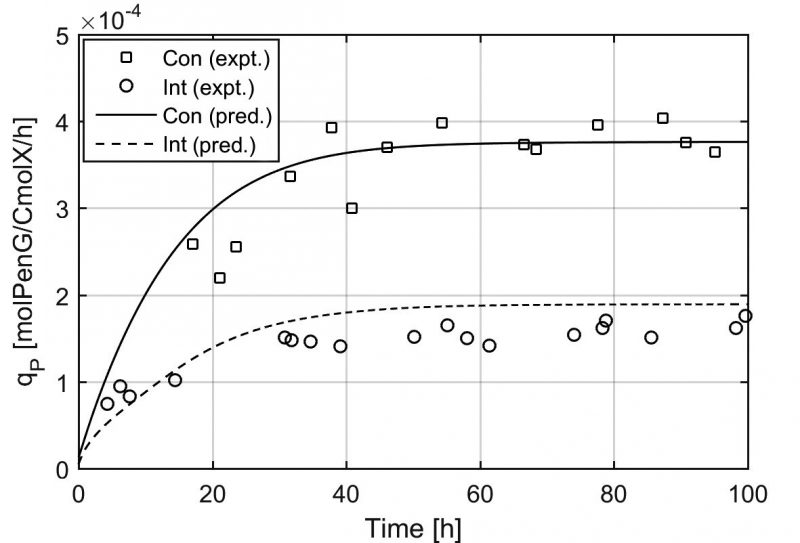
唐文俊等通過對產黃青霉開展多批次不同稀釋率下的恒化實驗、小時級的連續稀釋率變化實驗、秒級的補料速率變化實驗等獲取了大量的精確定量胞內代謝物濃度信息,利用同種類/同工特性建立“池”的概念,利用基礎元素守衡、電子及還原力守衡建立了一個具有5個代謝物池、4個相關調控酶池的結構化細胞代謝模型,并且為之建立了數十個相關動力學描述。最終的模型既能夠成功模擬小時級/天級的反應器內菌量增加、產物積累的過程,也能夠準確描述菌體在遭遇瞬時低糖濃度等不利條件下的應激響應,為與CFD計算流體力學的整合模擬提供了基礎。
原文鏈接:
http://onlinelibrary.wiley.com/doi/10.1002/bit.26294/full
DOI: 10.1002/bit.26294
原文摘要:
A powerful approach for the optimization of industrial bioprocesses is to perform detailed simulations integrating large scale computational fluid dynamics (CFD) and cellular reaction dynamics (CRD). However, complex metabolic kinetic models containing a large number of equations pose formidable challenges in CFD-CRD coupling and computation time afterward. This necessitates to formulate a relatively simple but yet representative model structure. Such a kinetic model should be able to reproduce metabolic responses for short-term (mixing time scale of tens of seconds) and long-term (fed-batch cultivation of hours/days) dynamics in industrial bioprocesses. In this paper, we used Penicillium chrysogenum as a model system and developed a metabolically structured kinetic model for growth and production. By lumping the most important intracellular metabolites in 5 pools and 4 intracellular enzyme pools, linked by 10 reactions, we succeeded in maintaining the model structure relatively simple, while providing informative insight into the state of the organism. The performance of this 9-pool model was validated with a periodic glucose feast-famine cycle experiment at the minute time scale. Comparison of this model and a reported black box model for this strain shows the necessity of employing a structured model under feast-famine conditions. This proposed model provides deeper insight into thein vivo kinetics and, most importantly, can be straightforwardly integrated into a computational fluid dynamic framework for simulating complete fermentation performance and cell population dynamics in large scale and small scale fermentors.




